IN-DEPTH: IN-situ DEtailed Phenotyping To High Resolution
Introduction
In the evolving field of biomarker discovery and spatial biology, IN-situ DEtailed Phenotyping To High Resolution (IN-DEPTH) represents a groundbreaking approach for high-dimensional cellular analysis. This method enables researchers to visualize and quantify proteomic and transcriptomic data within the native micro-environment, offering unparalleled insights into tissue heterogeneity and disease pathology.
What is IN-DEPTH?
IN-DEPTH is an advanced imaging and molecular profiling technique that integrates multiple high-resolution analytical platforms. It combines antibody-oligonucleotide conjugates, fluorescence imaging, and computational analysis to map cellular phenotypes with exceptional specificity and spatial accuracy.
Key Features of IN-DEPTH:
- Multiplexed Biomarker Detection: Enables simultaneous analysis of dozens to hundreds of proteins and gene expression signatures using a single tissue section.
- Preservation of Spatial Context: Maintains tissue architecture, allowing researchers to perform RNA and protein analysis.
- High Sensitivity & Resolution: Detects rare cell populations and subtle phenotypic variations.
- Integration with AI & Computational Tools: Facilitates large-scale data processing.
How Does IN-DEPTH Work?
The IN-DEPTH workflow involves several critical steps:
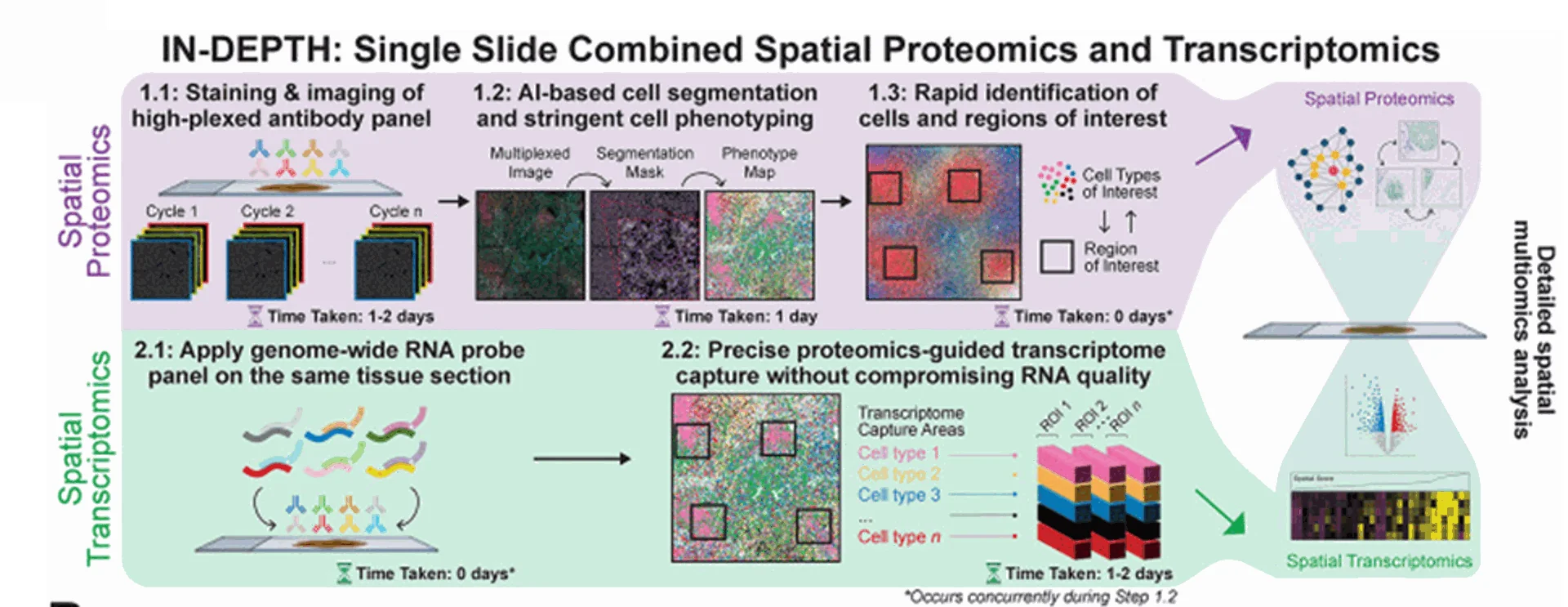
Image from Yeo et al 2024
1.0 Single cell spatial proteomic analysis
1.1 Tissue samples are fixed and permeabilized, then highly specific antibody-oligonucleotide conjugates (AOCs) are applied to target proteins of interest. High-plex immunofluorescence images are captured followed by repeat cycles using different antibody-oligo conjugates.
1.2 & 1.3 AI-driven segmentation and clustering algorithms identify cellular subtypes and functional states.
2.0 Transcriptomics
2.1 Genome wide RNA probes added to the same tissue section.
2.2 The proteome map generated in step 1.0, is used to capture high quality RNA data.
Applications of IN-DEPTH
The versatility of IN-DEPTH extends across multiple research and clinical domains:
- Cancer Research: Maps tumor microenvironment interactions and immune infiltration patterns.
- Neuroscience: Profiles neuronal subpopulations and synaptic connectivity.
- Immunology: Identifies immune cell subsets and their functional dynamics in tissue.
- Personalized Medicine: Aids in biomarker discovery for targeted therapy development.
Advantages Over Conventional Methods
Compared to traditional immunohistochemistry, IN-DEPTH offers:
- Higher Multiplexing Capacity: Detects more markers in a single sample.
- Better Spatial Resolution: Captures cellular interactions in tissue architecture.
- Improved Data Quantification: Provides precise molecular readouts.
Conclusion
As the demand for high-resolution, multi-omic spatial analysis grows, IN-DEPTH stands out as a cutting-edge solution for in situ phenotyping. Its ability to deliver rich, spatially resolved molecular insights makes it a powerful tool for both basic research and clinical applications.