Proximity Ligation Assays for Analyzing Protein - Protein Interactions.
Introduction to Proximity Ligation Assays (PLA)
Proximity Ligation Assay (PLA) is a highly sensitive molecular biology technique designed to detect and visualize protein-protein interactions directly within cells and tissues. Unlike traditional methods such as co-immunoprecipitation or fluorescence resonance energy transfer (FRET), PLA offers spatial precision by identifying proteins that are in close proximity, typically less than 40 nanometers apart. This allows researchers to study biological interactions in their natural cellular environment with high specificity. PLA has become an essential tool in biomedical research, shedding light on disease mechanisms, intracellular signaling pathways, and potential therapeutic targets.
How PLA Works: Detecting Protein Interactions In Situ
The core principle of PLA involves converting protein interactions into detectable DNA signals. It begins with two primary antibodies, each specific to one of the proteins of interest, conjugated to unique DNA oligos. When the target proteins are near each other, the attached oligos are brought into close proximity and ligated to form a circular DNA molecule. This DNA circle then undergoes rolling circle amplification (RCA), generating a long single-stranded DNA product. Fluorescently labeled probes complementary to this DNA are then used to visualize the interaction as bright, discrete spots under a fluorescence microscope.
In PLA, various assay formats have been developed to accommodate different experimental needs and levels of detection sensitivity. One common approach uses directly labeled primary antibodies, where each antibody specific to a target epitope is conjugated with a unique oligo. This format reduces background by minimizing the number of binding steps, making it ideal for high-specificity applications where well-characterized primary antibodies are available. Alternatively, the indirect PLA format uses unlabeled primary antibodies followed by species-specific secondary antibodies conjugated to oligos. This format is highly flexible, allowing for a broader selection of validated primary antibodies, and is particularly useful when the direct conjugation of oligonucleotides to primary antibodies is impractical.
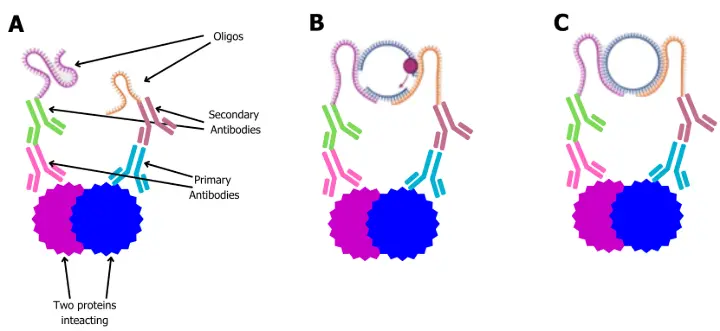
A - primary antibodies bind to the proteins of interest. Oligo-conjugated secondaries specifically interact with the relevant primary antibodies.
B - When the oligos are in close proximity, connector DNA strands join and become ligated in the presence of ligase, creating a close circular DNA loop (C).

D - rolling circle amplification extends the DNA sequence.
E - fluorescently labeled oligos hybridize to the extended DNA sequence, ready to be analyzed or measured using standard fluorescent microscopy.
Advantages of Using PLA Over Traditional Methods
Proximity ligation assay (PLA) offers several compelling advantages over conventional techniques for studying protein–protein interactions, including co-immunoprecipitation (co-IP), FRET, and pull-down assays. One of PLA's most notable strengths is its exceptional sensitivity, enabling the detection of transient or weak interactions that are often missed by methods like co-IP, which typically require stable protein complexes to withstand lysis and washing steps. While co-IP is a widely used and well-established technique, it lacks spatial resolution and often necessitates large amounts of starting material, which limits its application in rare cell types or precious clinical specimens. Moreover, co-IP is performed in lysates, meaning that any subcellular localization of the interaction is lost. In contrast, PLA is performed in situ within fixed cells or tissues, preserving the native cellular context and allowing researchers to pinpoint the exact subcellular location of interactions. Additionally, PLA does not require overexpression, epitope tagging, or protein purification, and instead detects endogenous proteins, offering more physiologically relevant insights. The incorporation of oligos in PLA further enhances signal strength, yielding high signal-to-noise ratios and enabling confident detection even at low protein abundance. Collectively, these advantages make PLA a powerful tool for studying protein interactions in their native cellular environment, especially in settings where conventional techniques fall short.
Applications of PLA in Biomedical Research
PLA is widely adopted in various areas of biomedical science, offering researchers a robust tool to unravel complex biological processes. In cancer research, PLA helps visualize interactions within signaling pathways and detect oncogenic modifications. Neuroscientists use PLA to study synaptic connections and protein aggregation in neurodegenerative disorders. In immunology, it enables the mapping of receptor-ligand interactions and signaling cascades. Additionally, PLA proves highly effective in analyzing formalin-fixed paraffin-embedded (FFPE) tissues, making it valuable for translational research and pathology.
Additional Materials and Reagents Needed for PLA
The enzymatic portion of the PLA workflow includes DNA ligase, which joins the oligonucleotides once they are brought into proximity, and DNA polymerase, which drives rolling circle amplification (RCA) to amplify the DNA signal tethered to the interaction site. Detection is then achieved using fluorescently labeled oligonucleotide probes that bind to the amplified DNA, creating discrete fluorescent spots visible under a fluorescence microscope.
In addition to the core reagents, blocking solutions and wash buffers are essential to minimize background and ensure specificity. Samples are typically prepared on microscopy-compatible glass slides, coverslips, or multi-well imaging plates, depending on the throughput and imaging platform.
Kits are available in ready-to-use commercial PLA formats, which offer tailored configurations for detecting protein–protein interactions, post-translational modifications, or even single-protein detection
Choosing the Right Antibodies for PLA
Antibody selection is a critical determinant of PLA success. For in-direct PLA formats, researchers must use high-affinity IgG-class antibodies, each raised in a different host species to ensure compatibility with the oligo-conjugated secondaries. Prior validation of these antibodies via standard immunofluorescence ensures effective target recognition. Equally important is confirming that the antibodies do not cross-react or bind non-specifically, which could generate false signals. In cases where appropriate antibodies are not available in different species, labeling the primary antibodies with carefully designed oligos make the assay adaptable to for a wide range of targets.
Hints and tips Performing a Successful PLA
1. Designing Oligonucleotides for PLA Probes
Designing effective oligonucleotides is essential for optimizing PLA performance. Ideal oligos range between 15 to 30 nucleotides. A balanced GC content (typically 40–60%) helps prevent undesirable secondary structures like hairpins or dimers. Sequence uniqueness must be verified using tools such as BLAST to ensure specificity. Additionally, chemical modifications—such as amino or biotin groups, must be incorporated for conjugation to antibodies. For RCA detection, separate primers and complementary fluorescent detection oligos are designed to recognize the amplified circle product.
2. Use Well-Validated Primary Antibodies
Choose primary antibodies that are highly specific and raised in different species. Cross-reactivity or poor affinity can result in background noise or weak signal.
3. Optimize Fixation Conditions
Fixation is critical for preserving native protein-protein interactions. Use fresh paraformaldehyde (typically 3.7–4%) and avoid over-fixation, which can mask epitopes and reduce probe accessibility.
4. Ensure Proper Permeabilization
Use detergents like Triton X-100 or Tween-20 to permeabilize cells without disrupting protein complexes. This step is essential for antibody and probe penetration.
5. Block Non-Specific Binding Sites
Apply a suitable blocking buffer (e.g., serum, BSA, or commercial blockers) to prevent non-specific binding of antibodies and PLA probes. This helps minimize background signal.
6. Titrate Antibody Concentrations
Optimize both primary and secondary (oligo-conjugated) antibody concentrations for your specific sample type. Too much antibody can increase background; too little reduces signal.
7. Include Rigorous Controls
Always run negative controls (e.g., single primary antibody, no primary antibody, or non-interacting protein targets) to assess background and confirm assay specificity.
8. Minimize Probe Cross-Talk
Avoid using PLA probes that might cross-react with unintended antibody species. Use well-matched oligos that are highly specific to the chosen secondaries.
9. Work Gently to Preserve Morphology
Handle cells and tissues carefully throughout the procedure to maintain cellular integrity. Harsh washes or mechanical force can distort morphology and displace interactions.
10. Use Fresh Enzymes and Buffers
Ligase, polymerase, and detection reagents should be fresh and stored properly. Degraded enzymes or expired buffers can severely impact ligation and amplification efficiency.
Conclusion
PLA is a powerful technique which has been commonly used to study protein-protein interactions. Interestingly, we are observing the assay being engineered to study protein-DNA and protein post-translational modifications as well. We have no doubt PLA assays will continue to evolve especially as antibody-oligo conjugates become more readily available.
References
- Proximity Ligation Assay (PLA) - Alam (2018)
- Proximity Ligation Assay for Detecting Protein-Protein Interactions and Protein Modifications in Cells and Tissues in Situ - Hegazy (2020)
- Nanoparticle-Based Proximity Ligation Assay for Ultra-Sensitive, Quantitative Detection of Protein Biomarkers - Chen (2019)